How To Draw Lineweaver Burk Plot
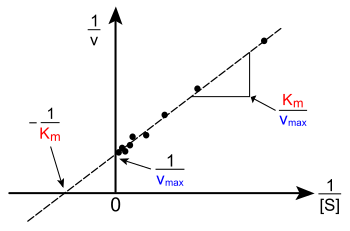
An example of a Lineweaver-Burk plot.
In biochemistry, the Lineweaver–Burk plot (or double reciprocal plot) is a graphical representation of the Lineweaver–Burk equation of enzyme kinetics, described past Hans Lineweaver and Dean Burk in 1934.[i] The Lineweaver–Burk plot for inhibited enzymes tin can be compared to no inhibitor to determine how the inhibitor is competing with the enzyme.[2]
The Lineweaver–Burk plot is correct when the enzyme kinetics obey platonic 2nd-club kinetics, however non-linear regression is needed for systems that do not behave ideally. The double reciprocal plot distorts the fault construction of the data, and is therefore not the nigh accurate tool for the determination of enzyme kinetic parameters.[3] Not-linear regression or culling linear forms of the Michaelis–Menten equation such every bit the Hanes-Woolf plot or Eadie–Hofstee plot are generally used for the calculation of parameters.[4]
Definitions for Interpreting Plot [edit]
: substrate concentration. The contained axis of a Lineweaver-Burk plot is the reciprocal of substrate concentration, .[2]
or : initial velocity of an enzyme-inhibited reaction. The dependent axis of the Lineweaver-Burk plot is the reciprocal of velocity, .[5]
: maximum velocity of the reaction. The y-intercept of the Lineweaver-Burk plot is the reciprocal of maximum velocity, .[two]
: the Michaelis abiding, a measure of enzyme analogousness. A lower means a higher affinity. Graphically, the x-intercept of the line is [v]
Gcat: turnover number, or reactions per unit time. The lower the Kcat the slower the reaction. 1000cat=5max/[Enzyme]. Graphically this can be evaluated by looking at Vmax. [2]
Catalytic Efficiency = One thousandtrue cat/KM. A fast catalyst and loftier affinity results in best catalytic efficiency.[5]
where is the concentration of inhibition and is the inhibitor abiding. Alpha determines the degree that bounden of an inhibitor furnishings enzyme kinetics of a substrate, it ever has a positive value.[5]
Derivation [edit]
The plot provides a very useful graphical method for analysis of the Michaelis–Menten equation, as it is hard to decide precisely the Vmax of an enzyme-catalysed reaction:
Taking the reciprocal gives united states:
The Lineweaver–Burk plot puts 1/[Due south] on the 10-axis and ane/V on the y-axis.[6]
Applications [edit]

Enzyme Inhibition displayed using Lineweaver-Burk (double reciprocal plots)
When used for determining the blazon of enzyme inhibition, the Lineweaver–Burk plot can distinguish competitive, pure non-competitive and uncompetitive inhibitors. The various modes of inhibition can be compared to the uninhibited reaction.
Competitive Inhibition [edit]
Vmax is unaffected by competitive inhibitors. Therefore competitive inhibitors have the aforementioned y-intercept as uninhibited enzymes (since V max is unaffected by competitive inhibitors the inverse of V max too doesn't change).
Competitive inhibition increases the GrandM, or lowers substrate analogousness. The KM inhibited is αKOne thousand.[v] Graphically this tin be seen as the inhibited enzyme having a larger ten-intercept.[2] The slopes of competitively inhibited enzymes and not-inhibited enzymes are different. Competitive inhibition is shown on the far left image.
Pure Noncompetitive Inhibition [edit]
With pure noncompetitive inhibition 5max is lowered with inhibition. Fivemax inhibited is αVmax.[5] This can be seen on the Lineweaver–Burk plot as an increased y-intercept with inhibition, every bit the reciprocal is plotted.[7]
Pure noncompetitive inhibition does non effect substrate analogousness, therefore KGrand remains unchanged. Graphically this tin can be seen in that enzymes with pure noncompetitive inhibition intersect with non-inhibited enzymes at the x-axis.[2] The slopes of pure noncompetitive inhibited enzymes and non-inhibited enzymes are different.[7] Pure noncompetitive inhibition is shown in the epitome far right image.
Mixed Inhibition [edit]
Pure noncompetitive inhibition is rare, meaning mixed inhibition is more probable to issue. In the case of mixed inhibition Vmax and MM are both effected at non-proportional rate. In well-nigh cases Vmax is decreased, while GChiliad is increased, meaning affinity unremarkably decreases with mixed inhibition. The lines of mixed inhibition and no inhibition intersect somewhere between the 10- axis and y- axis, simply never on an centrality with mixed inhibition.[5]
Uncompetitive Inhibition [edit]
Vmax decreases with uncompetitive inhibition. Fivemax inhibited is αVmax.[5] This tin be seen on the Lineweaver–Burk plot equally an increased y-intercept with inhibition, as the reciprocal is plotted.[7] This relationship is seen in both uncompetitive inhibition and pure competitive inhibition.[5]
Substrate affinity increases with uncompetitive inhibition, or lowers One thousandOne thousand. The inhibited K1000 is KM/α. Graphically this means that enzymes with uncompetitive inhibition will accept a smaller 10-intercept than non inhibited enzymes.[5] Despite the x-intercept and y-intercept of uncompetitive inhibition both irresolute, the slope remains constant. Graphically uncompetitive inhibition can be identified in that the line of inhibited enzyme is parallel to non-inhibited enzyme. Uncompetitive inhibition is shown in the middle image.
Bug with Lineweaver-Burk [edit]
While the Lineweaver-Burk is useful for determining important variables in enzyme kinetics, it is prone to error. The y-centrality of the plot takes the reciprocal of the rate of reaction, meaning minor errors in measurement are more noticeable.[8] Additionally, the values derived from depression [Southward] (and hence the more mistake decumbent values) are on the far right of the plot and have a larger impact on the slope of the line, and thus in particular on the value of ThouM.
See also [edit]
- Michaelis–Menten kinetics
- Eadie–Hofstee diagram
- Hanes–Woolf plot
References [edit]
- ^ Lineweaver, Hans; Burk, Dean (March 1934). "The Determination of Enzyme Dissociation Constants". Journal of the American Chemical Lodge. 56 (three): 658–666. doi:10.1021/ja01318a036. ISSN 0002-7863.
- ^ a b c d e f Ahern, Rajagopal (2013). "Biochemistry Costless and Easy". Biochemistry and Molecular Biology Education. 45 (i): 90–110. doi:ten.1002/bmb.20979. ISSN 1470-8175. PMID 27228905. S2CID 34758190.
- ^ Srinivasan, Bharath (18 March 2021). "Explicit Treatment of Non‐Michaelis‐Menten and Atypical Kinetics in Early on Drug Discovery**". ChemMedChem. xvi (six): 899–918. doi:ten.1002/cmdc.202000791. PMID 33231926. S2CID 227157473.
- ^ Greco, W. R.; Hakala, M. T. (1979-12-10). "Evaluation of methods for estimating the dissociation constant of tight binding enzyme inhibitors". The Periodical of Biological Chemistry. 254 (23): 12104–12109. doi:10.1016/S0021-9258(xix)86435-ix. ISSN 0021-9258. PMID 500698.
- ^ a b c d e f chiliad h i j Miesfeld, Roger L. (2017). Biochemistry. Megan Thou. McEvoy (1 ed.). New York, NY: W.W. Norton & Company. pp. 340–370. ISBN978-0-393-61402-2. OCLC 952277065.
- ^ Christensen, Siegfried B.; DeWolf, Walter E.; Ryan, Yard. Dominic; Torphy, Theodore J. (1996-01-01), Schudt, Christian; Dent, Gordon; Rabe, Klaus F. (eds.), "thirteen - Molecular Aspects of Inhibitor Interaction with PDE4", Phosphodiesterase Inhibitors, Handbook of Immunopharmacology, San Diego: Academic Press, pp. 185–207, doi:10.1016/b978-012210720-7/50015-0, ISBN978-0-12-210720-7 , retrieved 2020-12-xv
- ^ a b c Ahern, Rajagopal, Tan (2018). "Biochemistry Gratis for All". Biochemistry and Molecular Biology Pedagogy. 45 (1): 356–400. doi:10.1002/bmb.20979. ISSN 1470-8175. PMID 27228905. S2CID 34758190.
{{cite periodical}}: CS1 maint: multiple names: authors listing (link) - ^ Dowd, John E.; Riggs, Douglas S. (February 1965). "A Comparison of Estimates of Michaelis-Menten Kinetic Constants from Various Linear Transformations". Journal of Biological Chemistry. 240 (ii): 863–869. doi:x.1016/s0021-9258(17)45254-9. ISSN 0021-9258.
External links [edit]
- NIH guide, enzyme assay evolution and analysis
Source: https://en.wikipedia.org/wiki/Lineweaver%E2%80%93Burk_plot
Posted by: ruffinhiscambeste.blogspot.com

![[S]](https://wikimedia.org/api/rest_v1/media/math/render/svg/292bbb82029aa583c5d2ac5fa1d7e4fedf537d8b)
![{\frac {1}{[S]}}](https://wikimedia.org/api/rest_v1/media/math/render/svg/13eaed9b93d8ed8ac729f8dbb895987dbcc6597d)







![{\displaystyle \alpha =1+{\frac {[I]}{K_{I}}}}](https://wikimedia.org/api/rest_v1/media/math/render/svg/358c1362b1fb070d1020cc0e03007338bdd79af3)
![[I]](https://wikimedia.org/api/rest_v1/media/math/render/svg/5962300a54e8ce8b5761dac9a5fbbca450c2ce0f)

![V={\frac {V_{{\max }}[S]}{K_{m}+[S]}}](https://wikimedia.org/api/rest_v1/media/math/render/svg/8fa5b35e10661c7592ae5bc59429f1033a4acbf8)
![{1 \over V}={{K_{m}+[S]} \over V_{{\max }}[S]}={K_{m} \over V_{\max }}{1 \over [S]}+{1 \over V_{\max }}](https://wikimedia.org/api/rest_v1/media/math/render/svg/262e1440a8ad30a692b153178eabbf6e7f45d48f)

0 Response to "How To Draw Lineweaver Burk Plot"
Post a Comment